Structural variation reshapes population gene expression and trait variation in 2,105 Brassica napus accessions
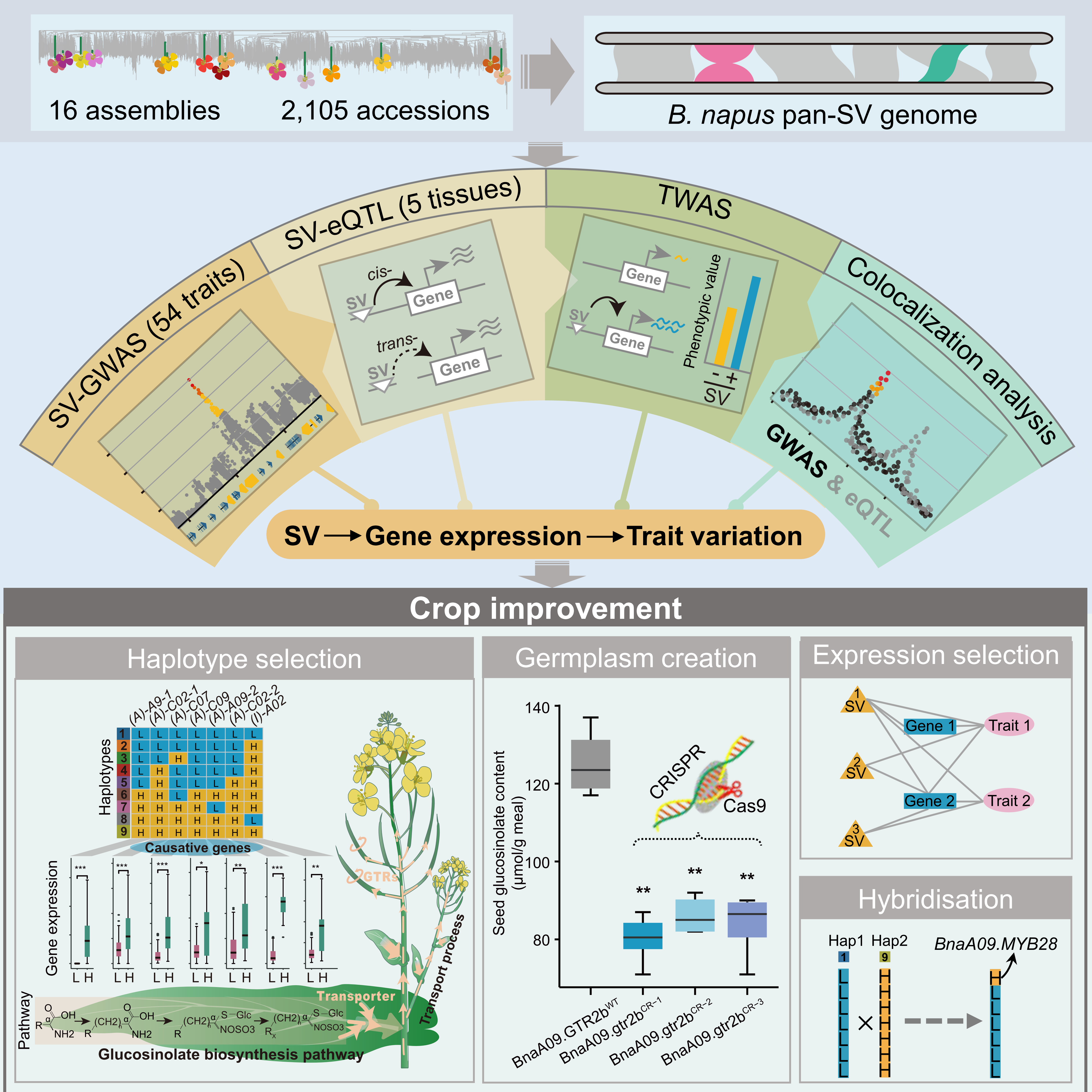
On 5 November 2024, the internationally authoritative academic journal Nature Genetics published online the joint research findings of the Oil Crops Research Institute of the Chinese Academy of Agricultural Sciences (OCRI-CAAS) and Huazhong Agricultural University (HZAU). The study reveals the patterns and mechanisms by which genomic structural variations (SVs) regulate gene expression and trait variation in B.napus. Furthermore, the team developed a novel high-throughput method for identifying SVs in polyploid genomes. This work provides new theoretical foundations and practical approaches for crop genomics research and genetic improvement.
SVs play a crucial role in phenotypic diversity and adaptive evolution. Understanding the relationship between SVs and agronomic traits, and applying this knowledge to crop improvement, is highly significant for discovering and creating elite germplasm resources, enhancing crop yield, improving quality, and boosting resistance. However, the abundance of repetitive sequences and complex variations within polyploid crop genomes have made large-scale SV detection inaccurate and costly. Consequently, systematic studies on how SVs regulate gene expression and trait formation at the species level have been lacking, creating a bottleneck for modern biological breeding.
To overcome this challenge, the Genomics and Disease Resistance Improvement Innovation Team at OCRI and the Oilseed Rape Team at HZAU conducted more than eight years of sustained collaborative research. They developed a novel high-throughput, high-precision method for identifying SVs in polyploid genomes. Applying this to polyploid B.napus, they comprehensively identified genome-wide SVs and revealed the extent and scale of SV impact on gene expression and thereby trait variation for the first time at the species level. This research not only provides a new strategy for the efficient large-scale identification and functional analysis of genetic variations, but also elucidated the mechanism coupling the biosynthesis and transport of glucosinolates – metabolites conferring pest and disease resistance in oilseed rape. Based on these findings, the team successfully created the elite oilseed rape germplasm resources with high resistance and superior quality by genome editing. These resources offer valuable genetic material for oilseed rape breeding programs. Overall, this study highlighting the impact of genome-wide and species-scale SVs provides a powerful methodological strategy and valuable resources for studying evolution, gene discovery, and breeding.
This research received support from the National Key Research and Development Program of China, the National Natural Science Foundation of China and the Agricultural Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences.
Link: https://www.nature.com/articles/s41588-024-01957-7

